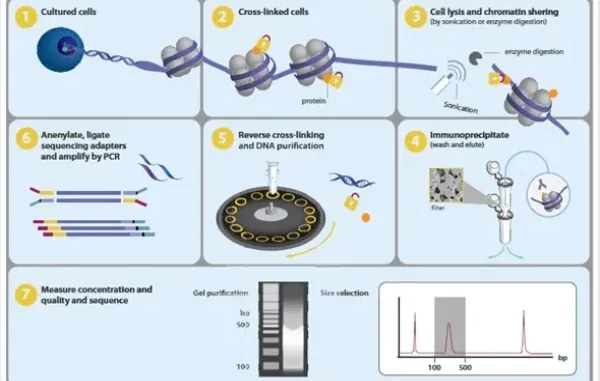
Chromatin immunoprecipitation (ChIP) followed by sequencing, known as ChIP-sequencing (ChIP-seq), is a powerful technique used to investigate protein-DNA interactions on a genome-wide scale. This method combines chromatin immunoprecipitation (ChIP) with high-throughput DNA sequencing to identify binding sites of DNA-associated proteins, such as transcription factors or specific histone modifications.
However, substantial variability persists due to experimental design differences, protocol execution, and reagent quality—all significantly affecting data comparability and biological interpretation. Recognizing these challenges, consortia like ENCODE and modENCODE have established comprehensive guidelines to enhance ChIP-seq quality. These guidelines define key parameters such as antibody validation standards, experimental replication strategies, and proper control implementation.
Additionally, commercial assay kits with pre-optimized reagents and standardized protocols help researchers achieve the consistency required for modern ChIP-seq workflows.
The role of assay kits in modern research
Assay kits are essential in molecular biology research, enhancing experimental speed, reproducibility, and standardization. This is especially essential for ChIP-seq, where variations impact data quality. High-quality kits address these issues via pre-optimized reagents and validated ChIP protocols, ensuring reliable results across labs. The ENCODE and modENCODE consortia emphasize antibody validation and experimental replication—requirements fulfilled by these kits. They include validated antibodies, aiding standardization. For ENCODE data submission, validation criteria specify that a histone band must constitute ≥50% of total signal in western blots.
Factors for selecting the right assay kit
Specificity: Opt for kits with validated data on key components, particularly antibodies. Off-target binding can lead to misleading results, highlighting the need for well-characterized reagents. In ENCODE studies, histone modification antibodies must capture at least 80% of the target modification in the immunoprecipitated histone signal, confirmed through mass spectrometry.
Sensitivity: The ability to detect low-abundance targets is important. While sequencing depth influences weak signal detection, initial immunoprecipitation quality (dependent on kit antibodies) determines ultimate sensitivity. ENCODE recommends ≥10 million uniquely mapped reads per replicate for point-source transcription factors.
Scalability: The evolving landscape of complex genomic analyses drives assay technology advancements. While sources do not detail commercial kit trends explicitly, ChIP-seq progress reveals parallel requirements. The growing capability to analyze hundreds of samples in parallel highlights a need for higher-throughput solutions, suggesting future demand for kits designed for large-scale analyses.
Emphasis on integrative analysis of multiple datasets points toward kits offering flexibility across conditions and targets, potentially incorporating multiplexing capabilities. The importance of quality metrics like FRiP and cross-correlation analyses suggests future kits may integrate quality control features. An analysis of published ChIP-seq data revealed only ~55% met high-quality criteria, underscoring standardization needs.
Key steps in ChIP-seq workflow
The successful execution of chromatin immunoprecipitation followed by sequencing (ChIP-seq) includes important steps:
- Cross-linking proteins to DNA
- Chromatin fragmentation (typically 150–500 bp, ideally 100–300 bp for most applications) using sonication or enzymatic digestion
- Target-specific immunoprecipitation
- Cross-link reversal
- DNA purification before sequencing
High-quality ChIP-seq relies on specific, high-affinity antibody selection and validation. ENCODE’s multi-tiered characterization begins with primary validation through immunoblot/immunofluorescence, requiring dominant specific bands of expected size. Secondary validation via target protein knockdown/knockout must show ≥50% ChIP signal reduction versus controls, confirming specificity. Researchers should prioritize well-validated antibodies from suppliers with robust quality control.
Appropriate controls remain essential for distinguishing biological signals from technical artifacts. Standard practice combines input DNA (processed without immunoprecipitation) and IgG controls (using non-nuclear antigen antibodies). ENCODE recommends sequencing controls to equal or greater depth than experimental samples for reliable enrichment identification. Unexpected control enrichment patterns may require careful interpretation.
Optimal outcomes require attention to the following key aspects:
- Comprehensive controls: Process input and IgG controls in parallel, sequencing to equivalent or greater depth.
- Biological replication: Minimum two independent replicates from distinct cell preparations ensure reproducibility.
- Quality metrics: Post-sequencing evaluation should include FRiP scores and cross-correlation analysis (normalized strand cross-correlation coefficient (NSC) >1.05, relative strand cross-correlation coefficient (RSC) >0.8). Subthreshold values indicate potential issues.
- Peak identification: Employ established algorithms with optimized parameters to reduce false positives.
By adhering to these practices—particularly using validated antibodies, proper controls, and thorough quality assessment—researchers can significantly enhance ChIP-seq reliability and biological interpretations.
Synergy between tools
The full potential of ChIP-seq technology emerges through the strategic integration of complementary tools. Rigorous antibody validation using techniques like immunoblotting (requiring ≥50% specific histone band signal) ensures reliable target enrichment. Adequate sequencing depth—ENCODE recommends ≥10 million uniquely mapped reads per replicate for mammalian transcription factors—guarantees comprehensive genomic coverage. Reproducibility assessment via irreproducible discovery rate (IDR) analysis with 1% thresholds provides quantitative consistency measures.
Specialized peak-calling software (eg, MACS) enables precise enriched region identification, while ENCODE guidelines create essential experimental frameworks. High-performance DNA purification kits optimize sequencing success. Advanced integrative tools like ChromHMM and Segway facilitate sophisticated genome annotation from ChIP-seq data. Comprehensive antibody characterization services ensure optimal reagent selection for specific experimental needs.
Conclusion
Modern ChIP-seq protocols, combined with precision-engineered assay kits, have revolutionized epigenomic research by enabling accurate, genome-wide mapping of protein-DNA interactions and histone modification patterns. This technological synergy provides essential insights into regulatory mechanisms governing cellular identity, developmental processes, and disease states. Quantitative quality metrics like IDR analysis offer researchers unprecedented confidence in their data interpretations. To advance your research capabilities, explore our portfolio of high-performance assay kits and leverage optimized ChIP-seq protocols to uncover a deeper understanding of genomic regulation.
Leave a Reply